
 |
Overview of the MAGE-TAB format |
|
On this page: See also: |
These notes provide an introduction to the MAGE-TAB format and a brief primer on how to submit microarray data to ArrayExpress in this format. MAGE-TAB is the successor to the Tab2MAGE format, developed by members of the microarray research community to extend the capabilities of the spreadsheet format. A full MAGE-TAB specification document is available from the MGED web site.
ArrayExpress supports submission of MAGE-TAB documents via a web-based submission system. This system allows the user to create an experiment submission and download an automatically-generated template MAGE-TAB document. This document can then be completed by the user and uploaded to the web page alongside their data files. For more information, please see the ArrayExpress submission help notes
[ Back ][ Top of page ]
The MAGE-TAB format uses a number of different files to capture information about a microarray experiment:
The IDF file is used to give an overview of the experiment, including the experimental variables (factors) used, protocols, quality control strategy, publication information and contact details. Also included in the IDF file is an (optional) list of sources from which controlled vocabulary terms may have been used elsewhere in the MAGE-TAB document. These term sources may be fully-fledged ontologies (e.g. the MGED ontology), databases providing queryable accession numbers (e.g. ArrayExpress), or simply a file defining terms for local users.
The SDRF file describes the relationship between every step in the chain of biological materials used in the experiment through to the hybridization, and the acquisition and normalization of data. It is similar in concept to the Hybridization section found in Tab2MAGE spreadsheets. Experimental factors, protocols, protocol parameters and term sources defined in the IDF are referenced by the SDRF.
The ADF file provides the array-level annotation for the experiment, relating the row-level identifiers in the data files to biological sequence annotation. Array designs are usually deposited in ArrayExpress as separate submissions to the experimental data, and in the case of commercial arrays may not need to be submitted to ArrayExpress at all.
Currently, ArrayExpress only supports submission of array designs using ADF files via the MIAMExpress web submission system. For help with this process, please see the MIAMExpress ADF help documentation. This array design component of MAGE-TAB will not be discussed further here.
An experimental data submission will usually consist of an IDF file, an SDRF file, and a series of data files. Typically there will be one raw data file per hybridization. Each hybridization may also have a normalized data file, or the final transformed data may be combined into a data matrix file. Note that the ArrayExpress MAGE-TAB generating system combines both the IDF and the SDRF into a single file format, for the sake of convenience.
For more detail on the MAGE-TAB document format, please see the MAGE-TAB specification document, available from the MGED web site.
[ Back ][ Top of page ]
The IDF component of a MAGE-TAB document consists of a set of unique tags attached to their corresponding values in a simple tab-delimited text format. For example, "Experiment Description" should be followed by a free-text description of the experiment. Most of the fields in the IDF document can handle multiple values. A full description of every valid IDF tag is given in the detailed IDF help notes. An example of an IDF document is given below:
| Investigation Title | Invasive vs. non-invasive strains of yeast | |
| Experimental Design | individual_genetic_characteristics_design | growth_condition_design |
| Experimental Factor Name | EF_Genotype | EF_GrowthCond |
| Experimental Factor Type | genotype | growth_condition |
| Person Last Name | Falstaff | Shakespeare |
| Person First Name | John | Bill |
| Person Email | jfalstaff@wagglespike.com | bills@wagglespike.com |
| Person Address | Ontario, Canada | Ontario, Canada |
| Person Affiliation | Windsor Laboratories | Windsor Laboratories |
| Person Roles | submitter;investigator | investigator |
| Quality Control Type | dye_swap_quality_control | |
| Public Release Date | 2004-08-30 | |
| PubMed ID | 12345678 | 87654321 |
| Publication Author List | Falstaff, J. and Shakespeare, B. | Goodfellow, R. et al. |
| Publication Title | Improved yeast flocculation | Yeast and beer: a retrospective |
| Publication Status | in preparation | submitted |
| Experiment Description | An experiment was performed to... | |
| Protocol Name | Yeast Growth | RNA extraction |
| Protocol Type | grow | nucleic_acid_extraction |
| Protocol Description | S. cerevisiae cultures were grown on... | Total cellular RNA was extracted... |
| Protocol Parameters | carbon source;temperature | |
| SDRF File | my_sdrf_file.txt |
Note that this is only a minimal subset of the available IDF tags. Blank lines may be included for legibility. Lines beginning with the "#" symbol are treated as comments and ignored. A full listing of all supported IDF tags can be found in these IDF help notes.
[ Back ][ Top of page ]
The SDRF file consists of a table in which each hybridization channel is represented by a row, and columns represent the steps of the experiment. In contrast to the Tab2MAGE format, the ordering of these columns is important, and should read left-to-right in chronological order. The overall organization of this table is shown below. To get more detail on the properties of each section, click on the relevant box below or read these detailed SDRF notes.
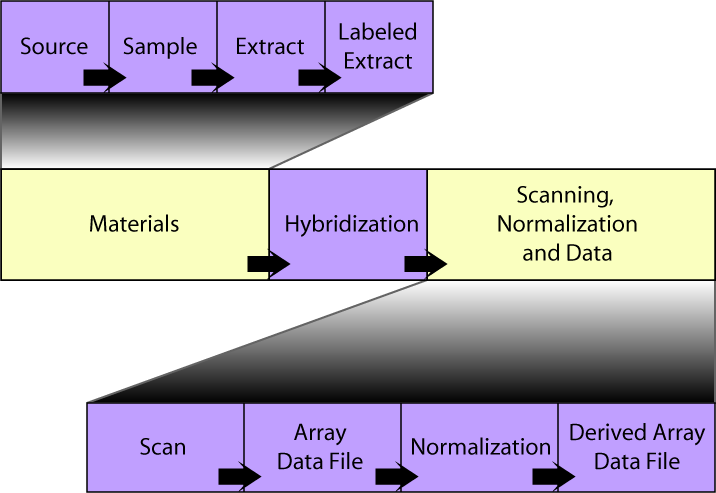
Each block in the diagram above starts with a "Name" or "File" column (e.g. "Extract Name", "Array Data File"), followed by a set of attribute columns. Each block is separated from its predecessor by "Protocol REF" columns containing references to the "Protocol Name" values defined in the IDF.
A further set of columns is used to specify the values for the variables ("experimental factors") within the experiment. These Factor Value[] columns reference the Experimental Factor Names defined in the IDF, and should be placed after the hybridization section (i.e., to the right of it, in or after the scanning, normalization and data section in the image above). The contents of these columns will usually duplicate those in a material Characteristics or a protocol Parameter Value column. See below for an example.
Below is a very simple example SDRF showing the links between materials and hybridization for a simple Affymetrix-based experiment:
| Source Name | Characteristics [OrganismPart] | Labeled Extract Name | Label | Protocol REF | Hybridization Name | Array Design REF |
| liver sample 1 | liver | LE 1 | biotin | Hyb protocol name | Hyb 1 | A-AFFY-33 |
| liver sample 2 | liver | LE 2 | biotin | Hyb protocol name | Hyb 2 | A-AFFY-33 |
| kidney sample 1 | kidney | LE 3 | biotin | Hyb protocol name | Hyb 3 | A-AFFY-33 |
| kidney sample 2 | kidney | LE 4 | biotin | Hyb protocol name | Hyb 4 | A-AFFY-33 |
Note that normally, many more "Characteristics[]" columns would be used to fully describe the Source. The SDRF might then continue in the following fashion, linking the Hybridization to data files and experimental factor values:
| Hybridization Name | Array Design REF | Scan Name | Array Data File | Protocol REF | Normalization Name | Derived Array Data File | Factor Value [FactorOP] |
| Hyb 1 | A-AFFY-33 | Scan 1 | Data1.CEL | Norm protocol name | Norm 1 | Data1.CHP | liver |
| Hyb 2 | A-AFFY-33 | Scan 2 | Data2.CEL | Norm protocol name | Norm 2 | Data2.CHP | liver |
| Hyb 3 | A-AFFY-33 | Scan 3 | Data3.CEL | Norm protocol name | Norm 3 | Data3.CHP | kidney |
| Hyb 4 | A-AFFY-33 | Scan 4 | Data4.CEL | Norm protocol name | Norm 4 | Data4.CHP | kidney |
In this case, the "Factor Value [FactorOP]" column refers to an Experimental Factor named "FactorOP", with Type "organism_part", from the IDF which would accompany this SDRF. Blank lines may be included for legibility. Lines beginning with the "#" symbol are treated as comments and ignored. Note that the examples on this page only illustrate a very minimal set of the available SDRF columns available. A full listing of all supported column names can be found in these SDRF help notes.
[ Back ][ Top of page ]
A variety of data file formats, produced by several different scanner makes and models, are supported by the ArrayExpress MAGE-TAB parser. A full list of supported formats can be found in the Tab2MAGE data file documentation. Note that the MAGE-TAB specification introduces a new file format, that of the data matrix, for files which contain data from multiple hybridizations. This new format is discussed below.
[ Back ][ Top of page ]
If you wish to represent data from more than one hybridization, scan or normalization in a single data file, you will need to reformat it as a MAGE-TAB Data Matrix. This is a simplified format which allows data columns to be mapped to rows in the SDRF file. The first header line of a Data Matrix file describes this mapping, and the second lists the quantitation types for each column (e.g. "log2 ratio"). The first column is used to map the data rows to identifiers from the array design used. Examples are shown here:
| Hybridization REF | Hyb1 | Hyb2 | Hyb3 | Hyb4 | Hyb5 | Hyb6 |
| Reporter REF | log2 ratio | log2 ratio | log2 ratio | log2 ratio | log2 ratio | log2 ratio |
| Probe 1 | 0.27 | 0.43 | 0.32 | 0.12 | 0.54 | 0.28 |
| Probe 2 | 1.67 | 1.46 | 1.91 | 1.49 | 1.50 | 1.89 |
| Probe 3 | 0.78 | 0.69 | 0.91 | 0.99 | 0.75 | 0.80 |
In this example, six hybridizations from the SDRF (Hyb1 - Hyb6) are being mapped to log2 ratio values. Each row of data is mapped to a Reporter Identifier defined by the array design (itself indicated in the SDRF file).
| Hybridization REF | Hyb1 | Hyb1 | Hyb2 | Hyb2 | Hyb3 | Hyb3 |
| CompositeElement REF | CELIntensity | CELStdev | CELIntensity | CELStdev | CELIntensity | CELStdev |
| Gene 1 | 22287.9 | 56.8 | 3222.9 | 111.1 | 9984.3 | 34.8 |
| Gene 2 | 267.4 | 7.6 | 118.1 | 7.6 | 236.8 | 9.0 |
| Gene 3 | 876.5 | 16.7 | 936.8 | 14.9 | 735.6 | 8.0 |
In this example, three hybridizations from the SDRF file (Hyb1, Hyb2 and Hyb3) are being mapped to data with two different quantitation types (CELIntensity, CELStdev). Each row of data is mapped to a CompositeElement Identifier (equivalent to MAGE CompositeSequence Identifiers) defined by the array design.
There are some limitations imposed by ArrayExpress when submitting data in this format. Firstly, each data matrix should correspond to hybridizations performed on a single array design. Experiments using multiple array designs should use one data matrix per design. Secondly, we rely on there being an ordered and regular organisation of the columns: first by hybridization, and then by quantitation type:
| Hybridization REF | Hyb1 | Hyb1 | Hyb2 | Hyb2 |
| Reporter REF | QT X | QT Y | QT X | QT Y |
| Hybridization REF | Hyb1 | Hyb2 | Hyb1 | Hyb2 |
| Reporter REF | QT X | QT X | QT Y | QT Y |
If your processed data does not readily fall into such a structure you may need to break the data up into multiple data matrices, and use the SDRF file to represent the relationships between them.
[ Back ][ Top of page ]
Please see the MAGE-TAB specification for further information and examples.