 |
SDRF help notes |
This page provides detailed help on the Sample and Data Relationship Format (SDRF), and lists all the available tags for use in MAGE-TAB SDRF documents. For an overview of the MAGE-TAB submission process, and examples of both IDF and SDRF, please see these submission help notes and MAGE-TAB overview. For more detail on the MAGE-TAB format, please see the MAGE-TAB specification.
MAGE-TAB documents may optionally include information on the sources of any controlled terms used, providing users with the ability to link to ontologies, databases or other sources of controlled vocabularies to describe their experiment. These so-called "Term Sources" are defined in the IDF and may be used throughout the document. If they are not used, then all controlled vocabulary terms are assumed to be user-defined.
[ Back ][ Top of page ]
The SDRF file consists of a table in which each hybridization channel is represented by a row, and columns represent the steps of the experiment. In contrast to the Tab2MAGE format, the ordering of these columns is important, and should read left-to-right in chronological order. The overall organization of this table is shown below. To get more detail on the properties of each section, click on the relevant box:
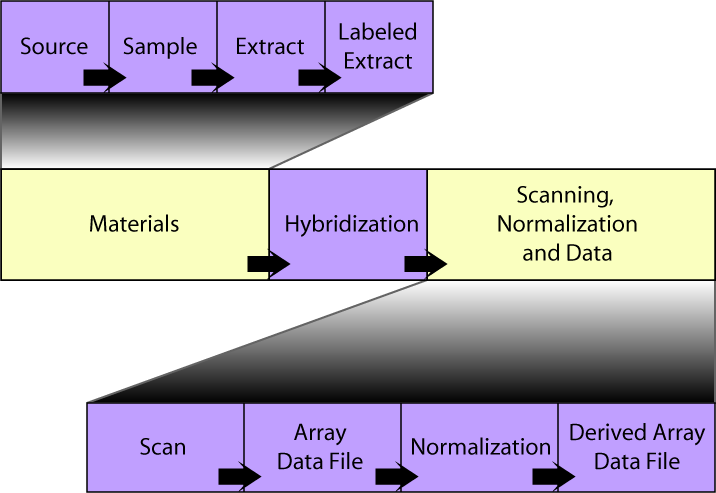
An experiment can be described in terms of a graph, in which the graph nodes correspond to materials or data files, and the graph edges (or arcs) correspond to treatments. Each block in the diagram above would be represented as a node in such a graph, with the treatments (protocols) acting as edges. Each node block starts with a "Name" or "File" column (e.g. "Extract Name", "Array Data File") identifying the type of node, followed by a set of attribute columns. Each block is separated from its predecessor by "Protocol REF" graph edge columns containing references to the "Protocol Name" values defined in the IDF.
A further set of columns is used to specify the values for the variables ("experimental factors") within the experiment. These Factor Value[] columns reference the Experimental Factor Names defined in the IDF, and should be placed after the hybridization section (i.e., to the right of it, in or after the scanning, normalization and data section in the image above). The contents of these columns will usually duplicate those in a material Characteristics or a protocol Parameter Value column. See below for an example.
[ Back ][ Top of page ]
Protocols
In most cases, each treatment
within an experiment will be represented simply by a Protocol REF column containing
references to the Protocol Names defined in the
IDF. If multiple protocols need to be chained together this can
be achieved using multiple Protocol REF columns.
Additionally, any Protocol Parameters associated with the protocol (as defined in the IDF) should have their values listed after the Protocol REF column. For example, with this in your IDF:
| Protocol Name | My Ext. Protocol | My Labeling Protocol |
| Protocol Type | nucleic_acid_extraction | labeling |
| Protocol Parameters | amplification;RNA quality | amount of RNA used |
The SDRF describing the use of these protocols might look as follows:
| Protocol REF | Parameter Value [RNA quality] | Parameter Value [amplification] | Protocol REF | Parameter Value [amount of RNA used] | Unit [MassUnit] |
| My Ext. Protocol | RIN 8 | RNA polymerase | My Labeling Protocol | 10 | ug |
Other columns which may be used to annotate these Protocol REF columns are: Performer, Date, and Comment.
Sources
Sources are the starting
material for the experiment. The section starts with a Source
Name column, which will typically be followed by several Characteristics columns and a Material Type column:
| Source Name | Material Type | Characteristics [Organism] | Characteristics [OrganismPart] | Characteristics [DiseaseState] | Characteristics [BioSourceType] |
| Tumor 1 | organism_part | Homo sapiens | mammary gland | invasive ductal carcinoma | frozen_sample |
Additional columns which may be used to annotate Sources are: Provider, Description, and Comment.
Samples
Samples represent steps in the
chain of treatments applied to the original Source. MAGE-TAB
allows you to create as many Sample steps as necessary:
| Source Name | Protocol REF | Sample Name | Material Type | Protocol REF | Sample Name | Material Type | Characteristics [OrganismPart] |
| Young Rat 99 | My growth protocol | Adult Rat 99 | whole_organism | My dissect protocol | Adult Rat Liver 99 | organism_part | liver |
For ArrayExpress submissions, typically only one or two Sample steps are needed. Columns which may be used to annotate Samples are: Characteristics[], Material Type, Description, and Comment.
Extracts
Extracts refer to the
extracted nucleic acid used in the experiment. Again, as many
Extract steps may be used as are necessary. For example, if you
need to represent separate nucleic acid extraction and chromatin
immunoprecipitation steps in your SDRF, we recommend that you
use two Extract steps. In most cases, however, a single Extract
Name column would suffice:
| Sample Name | Material Type | Protocol REF | Extract Name | Material Type |
| Rat Liver 99 | organism_part | My Ext. Protocol | Liver RNA 99 | total_RNA |
Columns which may be used to annotate Extracts are: Characteristics[], Material Type, Description, and Comment.
Labeled Extracts
The Labeled Extracts
in an experiment are those materials which have been conjugated
to a label of some kind, prior to hybridization on an
array. Typically there is only one Labeled Extract step. For
submission to ArrayExpress, a Label column
must be included with the Labeled Extract Name
column to indicate which label (and therefore scanner channel)
corresponds to which sample:
| Extract Name | Material Type | Protocol REF | Labeled Extract Name | Label | Material Type |
| Liver RNA 99 | total_RNA | My labeling protocol | Liver LE 99 Cy3 | Cy3 | synthetic_DNA |
| Kidney RNA 34 | total_RNA | My labeling protocol | Kidney LE 34 Cy5 | Cy5 | synthetic_DNA |
Note that it it wise to also include the Label in the Labeled Extract Name itself, so that unique objects are correctly created for each labeled extract. Columns which may be used to annotate Labeled Extracts are: Characteristics[], Material Type, Description, and Comment.
Hybridizations
The hybridization of
Labeled Extract to an array is a key step in the SDRF, since it
connects the "materials" section of the SDRF from the "data"
section. For submission to ArrayExpress, an Array Design REF column
must be included with the Hybridization Name column,
indicating which array design was used in the hybridization:
| Labeled Extract Name | Label | Hybridization Name | Array Design REF |
| Liver LE 1 | Cy3 | Liver vs. Kidney 1 | A-MEXP-88 |
| Kidney LE 1 | Cy5 | Liver vs. Kidney 1 | A-MEXP-88 |
It is also possible to use Comment columns to annotate both Hybridization Name and Array Design REF columns. Note that the values in Hybridization Name columns may be used in Data Matrix files to link columns of data to individual hybridizations.
Scans
If desired, the act of scanning
the hybridized array may be represented as a distinct node in
the experimental graph, and encoded in the SDRF using Scan Name columns. These columns are
optional, but can be useful in cases where e.g. multiple scans
have been made of a single hybridized array, but where the data
files do not explicitly reflect this:
| Hybridization Name | Array Design REF | Scan Name | Array Data File |
| Liver vs. Kidney 1 | A-MEXP-88 | LK1 First Scan | Data1.txt |
| Liver vs. Kidney 1 | A-MEXP-88 | LK1 Second Scan | Data1.txt |
Again, Comment columns may be used to further annotate Scan Name columns, where appropriate. Note that the values in Scan Name columns may be used in Data Matrix files to link columns of data to individual scanning events.
Array Data Files
The raw data files
generated by an investigation should be listed in an Array Data File column
following the Hybridization Name and
(optional) Scan Name columns:
| Hybridization Name | Array Design REF | Array Data File | Comment [EXP] |
| Liver vs. Kidney 1 | A-AFFY-33 | Data1.CEL | Data1.EXP |
Comment columns can be used to add information relating to Array Data Files. For example, if you are coding an Affymetrix-based experiment and you wish to include the EXP files in your submission, you should list them in a "Comment[EXP]" column following the "Array Data File" column, as shown above.
Normalizations
Similarly to the use of
Scan Name columns above, it is possible to represent the
act of normalizing your data independently from the listing of
data files themselves. This is done using the optional Normalization Name column:
| Hybridization Name | Array Design REF | Array Data File | Normalization Name | Derived Array Data File |
| Liver vs. Kidney 1 | A-AFFY-33 | Data1.CEL | Norm 1 | Data1.CHP |
Again, Comment columns may be used to further annotate Normalization Name columns, where appropriate. Note that the values in Normalization Name columns may be used in Data Matrix files to link columns of data to individual normalization events.
Derived Array Data Files
The processed
data files which have been derived from the raw data should be
listed in an Derived Array Data File
column. Note that this generally only applies to
processed data arranged into one file per hybridization (or
scan, or normalization). If your files contain processed data
columns for more than one hybridization, you should reformat
these into the MAGE-TAB
Data Matrix format and include them instead in a Derived Array Data Matrix File
column. Multiple steps of normalization can be captured:
| Hybridization Name | Array Design REF | Array Data File | Normalization Name | Derived Array Data File | Normalization Name | Derived Array Data Matrix File | Comment [CDF] |
| Liver vs. Kidney 1 | A-AFFY-33 | Data1.CEL | MAS5 Norm 1 | Data1.CHP | RMA Norm | RMANormData.txt | HG-U133A.cdf |
| Liver vs. Kidney 2 | A-AFFY-33 | Data2.CEL | MAS5 Norm 2 | Data2.CHP | RMA Norm | RMANormData.txt | HG-U133A.cdf |
| Liver vs. Kidney 3 | A-AFFY-33 | Data3.CEL | MAS5 Norm 3 | Data3.CHP | RMA Norm | RMANormData.txt | HG-U133A.cdf |
| Liver vs. Kidney 4 | A-AFFY-33 | Data4.CEL | MAS5 Norm 4 | Data4.CHP | RMA Norm | RMANormData.txt | HG-U133A.cdf |
In the above example, the columns from the "RMANormData.txt" data matrix file could be linked to either the Hybridization Names or the previous set of Normalization Names, allowing for flexible representation of the flow of data through the process. See the Data Matrix notes for details of how these links are encoded in the data matrix file header.
Comment columns may be used to add information relating to processed data files. For example, when coding an Affymetrix-based experiment with a Data Matrix file, as in the example above, a Comment[CDF] column should be used to indicate which Affymetrix library ("CDF") file applies to these data.
Factor Values
The Factor Values for an
experiment are the values of the variables under
investigation. For example, an experiment studying the effect of
different compounds on a cell culture would have "compound" as
an experimental variable. These variables are listed in the IDF
as "Experimental Factor Names" with associated Types:
| Experimental Factor Name | Cells | Drug |
| Experimental Factor Type | cell_line | compound |
| Protocol Name | All Treatments | |
| Protocol Parameters | drug compound |
Given the above definitions in the accompanying IDF, the SDRF file can then reference these factors when we come to list the factor values:
| Source Name | Characteristics [CellLine] | Protocol REF | Parameter Value [drug compound] | Hybridization Name | Factor Value [Cells] | Factor Value [Drug] |
| Line 1 | Jurkat | All Treatments | imatinib | Jurkat vs imatinib | Jurkat | imatinib |
| Line 1 | Jurkat | All Treatments | lapatinib | Jurkat vs lapatinib | Jurkat | lapatinib |
| Line 2 | RKO | All Treatments | imatinib | RKO vs imatinib | RKO | imatinib |
| Line 2 | RKO | All Treatments | lapatinib | RKO vs lapatinib | RKO | lapatinib |
Note that there is inevitably duplication between Factor Values and values entered elsewhere in the SDRF. It is particularly common to have the Factor Value column duplicate either a Characteristics[] column or a Parameter Value[] column.
Factor Value columns may be placed anywhere after the hybridization section of the SDRF, although they should be placed to avoid disrupting any of the other sections. This is most easily achieved by adding them at the end (i.e., the far right) of the SDRF.
[ Back ][ Top of page ]
Source
Name
Used as an identifier within
the MAGE-TAB document. This column contains user-defined names for the Source materials. The following
columns can be used to annotate Source Name columns:
Sample
Name
Used as an identifier within
the MAGE-TAB document. This column contains user-defined
names for each Sample
material. The following columns can be used to annotate Sample
Name columns:
Extract
Name
Used as an identifier within
the MAGE-TAB document. This column contains user-defined
names for each Extract
material. The following columns can be used to annotate Extract
Name columns:
Labeled Extract Name
Used as an identifier within the MAGE-TAB
document. This column contains user-defined names for
each Labeled Extract
material. The following columns can be used to annotate Labeled
Extract Name columns:
Hybridization Name
Used as an identifier within the MAGE-TAB
document.This column contains user-defined names for each
Hybridization. The
following columns can be used to annotate Hybridization
Name columns:
Assay
Name
Used as an identifier within
the MAGE-TAB document.This column contains user-defined
names for each Assay. "Assay Name" may be used instead of
"Hybridization Name" to identify generic biological assays, such
as rtPCR. Note that this column should not be used for
submission of regular microarray experiments to
ArrayExpress. The following columns can be used to annotate
Assay Name columns:
Note that as of MAGE-TAB version 1.1, all Assay Name columns must be followed by a Technology Type column.
Scan
Name
Used as an identifier within
the MAGE-TAB document. This optional column contains
user-defined names for each Scan
event. The following columns can be used to annotate Scan Name
columns:
Normalization Name
Used as an identifier within the MAGE-TAB
document. This optional column contains user-defined
names for each Normalization event. The
following columns can be used to annotate Normalization Name
columns:
Array
Data File
This column contains a list of raw data files, one for each
row of the SDRF file, linking these data files to their
respective hybridizations. The following columns can be used to
annotate Array Data File columns:
Derived Array Data File
This column
contains a list of processed data files,
one for each row of the SDRF file, linking these data files to
their respective hybridizations. The following columns can be
used to annotate Derived Array Data File columns:
Array Data Matrix File
This column
contains a list of raw data matrix files, where data from
multiple hybridizations is stored in a single file, and the data
mapped to each hybridization via the Data Matrix format
itself. The following columns can be used to annotate Array Data
Matrix File columns:
Derived Array Data Matrix File
This
column contains a list of processed data matrix files, where
data from multiple hybridizations is stored in a single file,
and the data mapped to each hybridization (or scan, or
normalization) via the Data Matrix format
itself. The following columns can be used to annotate Derived
Array Data Matrix File columns:
Image
File
This optional column contains a list of image
files, one for each row of the SDRF file, linking these image
files to their respective hybridizations. Note that ArrayExpress
does not store image data due to size constraints on the
database. If desired, you may use this column to include links
to image files stored on your local webserver. The following
columns can be used to annotate Derived Array Data File
columns:
Array
Design REF
This column contains references to the array
design used for each hybridization. For ArrayExpress submissions
this should be an ArrayExpress accession number,
e.g. "A-AFFY-33". The following columns can be used to annotate
Array Design REF columns:
The Term Source REF column here can be used to point to the source of the array design referenced; however for ArrayExpress submissions this should always be ArrayExpress itself, and so this column is in effect ignored.
Protocol
REF
This column contains references to Protocol Names
defined in the IDF, or accession numbers of protocols already
deposited with ArrayExpress. The following columns can be used
to annotate Protocol REF columns:
The Term Source REF column here can be used to point to the source of the protocol referenced, if it is not contained within the IDF; for ArrayExpress submissions this should always be ArrayExpress itself, and a suitable ArrayExpress Term Source should be defined in the IDF.
Characteristics[<category term>]
Controlled vocabulary term or
measurement. Used as an attribute column following Source Name, Sample Name, Extract Name, or Labeled Extract Name. This
column contains terms describing each material according to the
characteristics category indicated in the column header. For
example, a column headed "Characteristics[OrganismPart]" would
contain individual OrganismPart terms. These terms may be
user-defined (the default), from an external ontology source
(indicated using a Term Source REF column), or a measurement
(indicated using a Unit[] column).
Provider
Used as an attribute column
following Source Name. A free-text
string identifying the organization or person from which the
Source was obtained.
Material
Type
Controlled vocabulary
term. Used as an attribute column following Source Name, Sample Name, Extract Name, or Labeled Extract Name. This
column contains terms describing the type of each material. For
ArrayExpress submissions this term should be an instance of MaterialType
from the MGED Ontology. Examples: whole_organism,
organism_part, cell, total_RNA. The following columns can be
used to annotate Material Type columns:
The Term Source REF column in this case would point to the ontology (defined in the IDF) from which the Material Type terms are taken (the MGED Ontology in the example above).
Label
Controlled vocabulary term. Used as an
attribute column following Labeled Extract Name. The label
compound which is conjugated to an Extract to create the Labeled
Extract. For ArrayExpress submissions this term should be an
instance of LabelCompound
from the MGED Ontology. Examples: Cy3, Cy5, biotin,
alexa_546. The following columns can be used to annotate Label
columns:
The Term Source REF column in this case would point to the ontology (defined in the IDF) from which the Label terms are taken (the MGED Ontology in the example above).
Technology Type
Controlled vocabulary term. Used as an
attribute column following Assay
Name. This column contains terms describing the type of each
generic (non-hybridization) assay. Example: rtPCR. The following
columns can be used to annotate Technology Type columns:
The Term Source REF column in this case would point to the ontology (defined in the IDF) from which the Technology Type terms are taken.
Factor
Value[<experiment factor name>]
Controlled vocabulary term or measurement.
This column contains terms describing the experimental factor
values (i.e., variables) for each row of the SDRF. The
Experimental Factor Name to which it pertains (from the
accompanying IDF) should be indicated in the column heading. For
example, if you have this in your IDF:
| Experimental Factor Name | TissueEF |
| Experimental Factor Type | organism_part |
You could then use this factor in your SDRF (assuming you had also defined the "Mouse Anatomy" term source in your IDF):
| Factor Value[TissueEF] | Term Source REF |
| brain | Mouse Anatomy |
| kidney | Mouse Anatomy |
| liver | Mouse Anatomy |
| intestine | Mouse Anatomy |
| pancreas | Mouse Anatomy |
The terms in the column may be user-defined (the default), from an external ontology source (indicated using a Term Source REF column), or a measurement (indicated using a Unit[] column).
In the example above, the column terms would be treated as describing organism parts. For more precise control over the treatment of these terms, the optional form "Factor Value [] ()" is available, e.g. "Factor Value [growth condition EF] (Nutrients)".
Performer
Used as an attribute column
following Protocol REF. The name of
the researcher who carried out the protocol.
Date
Used as an attribute column following Protocol REF. The date (and time, where
available) upon which the protocol was performed, in the
following format: YYYY-MM-DD
Parameter Value[<protocol
parameter>]
Used as an attribute column following Protocol REF columns. This column
contains values for the protocol parameters referenced in the
column header. The following columns can be used to annotate
Parameter Value[] columns:
For example, if a Protocol Name "Array Hybridization" is defined in the accompanying IDF, with Protocol Parameters "hyb temp;hyb volume", the following would be valid:
| Protocol REF | Parameter Value [hyb temp] | Unit [TemperatureUnit] | Parameter Value [hyb volume] | Unit [VolumeUnit] |
| Array Hybridization | 55 | degrees_C | 100 | ul |
Unit[<unit
category>]
Controlled vocabulary
term. Used as an attribute column following Characteristics[], Factor Value[] or Parameter Value[]. This column
contains terms describing the unit(s) to be applied to the
values in the preceding column. The type of unit is included in
the column heading, e.g. "Unit[TimeUnit]". These unit types
should correspond to Unit
subclasses from the MGED Ontology. The following columns can
be used to annotate Unit[] columns:
The Term Source REF column in this case would point to the ontology (defined in the IDF) from which the Unit terms are taken.
Description
Used as an attribute column
following Source Name, Sample Name, Extract Name, or Labeled Extract Name. A
free-text description to be attached to the corresponding
material. To be used sparingly, if at all - most annotations
should be provided using controlled vocabulary terms, using Characteristics[] columns.
Term
Source REF
Used as an attribute column following any
controlled vocabulary column (e.g., Characteristics[], or column
allowing reference of external entities (e.g., Protocol REF. This column contains
references to ontology or database Term Sources defined in the
IDF, and from which the values in the previous column were
taken. The following columns can be used to annotate Term Source
REF columns:
Term Accession Number
Used as an
attribute column following Term
Source REF columns. This column contains the accession
numbers from the term source used to identify the ontology or
database terms in question. For example:
| Source Name | Characteristics [DiseaseState] | Term Source REF | Term Accession Number |
| Sample 1 | acute lymphocytic leukemia | NCI Metathesaurus | C0023449 |
(This example relies on the "NCI Metathesaurus" Term Source having been pre-defined in the IDF accompanying the SDRF.)
Comment[<comment name>]
This
column can be used to annotate the main graph node and edge
columns listed above. It is included as an extensibility
mechanism, and should not generally be used to encode meaningful
biological annotation. The column heading should contain a name
for the type of values included in the column.
[ Back ][ Top of page ]
Please see the MAGE-TAB specification for further information and examples.